|
To learn how tissues develop and maintain their organization — and
especially to learn what goes wrong when cancer strikes — it's essential
to study individual cells and their nuclei within tissues. The problem
is that in real tissues, and in many cell cultures grown in the laboratory,
cells are often tightly clustered; their boundaries and the borders of
their nuclei are hard to distinguish.
 |
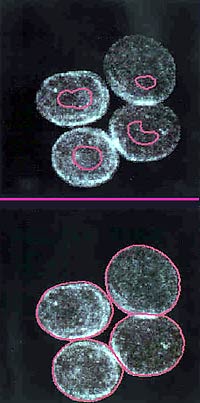
A COMPUTER PROGRAM AUTOMATICALLY PLANTS A "SEED" IN
THE IMAGE OF A NUCLEUS. THE PROGRAM THEN GROWS THE SEED UNTIL THE
ENVELOPE OF THE NUCLEUS IS OUTLINED.
|
Now Carlos Ortiz de Solorzano of Berkeley Lab's Life Sciences Division
and Ravi Malladi of the Lab's Computing Sciences organization, with their
colleagues Sophie Lelièvre and Stephen Lockett, have joined forces to
apply new biological and computational methods to mark and detect the
boundaries of closely packed cells and nuclei crowded together under the
microscope. Their methods work not only with the envelopes of nuclei but
with the membranes of tightly clustered cells as well.
To label nuclei the researchers first chose a set of stains that specifically
bind to the protein lining of the inner and outer layers of the nuclear
membrane, called lamina. Next they developed programs that could find
and outline the stained lamina in the microscopic images after they were
stored on the computer.
"Typically, researchers use fluorescent stains that bind to the
DNA inside the nucleus, not to the nuclear envelope," says Ortiz
de Solorzano. "But when nuclei are clumped together" —
typical in tumor tissue —"there's no contrasting background
area between them, and there's no easy way to distinguish one from the
next."
Seen through the microscope, even stained nuclear membranes may present
a mixed bag of dim and cluttered outlines, and while the human eye can
pick through these one by one, the new program identifies each nucleus
and marks it with a tiny internal graphical "seed," which grows
until its periphery corresponds to the outline of the lamina.
"Planting just one seed in each nucleus is important, " says
Ortiz de Solorzano. "If no seed is planted, the algorithm will be
unable to find the boundaries, but if more than one seed is planted, they
will mistakenly divide the nucleus into more than one object."
Malladi explains that the process begins by automatically choosing seed
points to lie inside the nuclei. This is done by computing a crude estimate
of the gradient magnitude and direction — a measure of change in
the intensity of the image's pixels — in the entire image and translating
the gradient points along the "inward" direction. As a result,
the gradient points corresponding to the nuclei boundaries reinforce each
other in a small region inside nuclei.
These ensembles of reinforced gradient points define the initial seed
points. "The gradients won't clump exactly in the center," Malladi
says, "but that doesn't matter as long as they are inside the nucleus."
Next the program moves the periphery of each seed (called the "front")
outward until it conforms to the shape defined by the stained lamina.
The movement proceeds in stages that are highly sensitive to changes in
pixel intensity; the front moves outward freely where changes are minor,
but slows almost to a stop when pixel intensity changes markedly, alerting
the program to a boundary.
| |
 |
| |
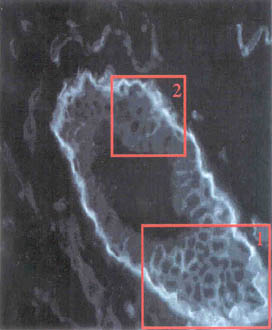
IN THIS IMAGE OF MAMMARY TISSUE FROM
A MOUSE, SOME CELLS ARE SO DENSELY CLUSTERED THEIR BOUNDARIES ARE
AMBIGUOUS. |
Having marked the inner membrane surface, the front is now allowed to
move beyond it, coming to a halt only when it encounters other boundaries
expanding from other nuclei. Then the front backtracks, seeking maximum
intensity values that clearly identify the stained outer lamina.
The algorithm works in similar ways to unambiguously outline whole cells
using, for example, fluorescently labeled integrins, which are proteins
specific to cell membranes.
In both cases "the program makes use of edge-finding algorithms
earlier developed for medical imaging," Malladi explains. "We
have also developed features that enable the program to find boundaries
accurately in very noisy images and to 'see' the right shape even where
there are discontinuities." These robust algorithms produce few errors
unless the original images are so bad that they are not worth using to
begin with.
|
|
| AFTER
SEEDS ARE PLANTED "INTERACTIVELY" (BY HUMAN EYE), THE NEW
PROGRAM OUTLINES THE CELL MEMBRANES CLEARLY. |
Computer scientist Malladi is excited about widening the frontiers of software
that can abstract visual information from a variety of disparate image sources,
ranging from medical images of bones and organs to problems in combustion
and fluid mechanics to the underground structures of oil reservoirs. Ortiz
de Solorzano, whose focus is biological image analysis, welcomes the nuclei
and cell detection techniques as a new means of "quantifying information
to understand biological processes."
The new techniques are described in the article "Segmentation of
nuclei and cells using membrane related protein makers," by C. Ortiz
de Solorzano, R. Malladi, S.A. Lelièvre, and S.J. Lockett, which appeared
in the March, 2001 issue of the Journal of Microscopy. Lockett
and Lelièvre, formerly of the Life Sciences Division, have recently accepted
positions outside Berkeley Lab.
Additional information:
|


